SDM Lab 8: Define a graphical DSML using EuGENia (updated 15/11/18)
In this lab, you'll create a graphical DSML and an editor for it. It will
feel like a complicated process the first time, but at the end, look at how
little you've actually had to write... We'll use the EuGENia toolset, which
seems to be reasonably robust. (You might like to look around afterwards
and see what other tool options are available.)
You can use either the Eclipse Modeling Edition or the specific Papyrus
distribution. Ways to install missing software, menu structure etc. differ
a little... these instructions were written using the former.
Getting started: create an ecore model for Paleo
You'll need a metamodel in ecore for the Paleo diet-food-classification
language. Since it's very simple you could just create one from scratch,
but let's illustrate how we can create an ecore model from a UML model,
just for fun...
Create or find a UML class diagram built in Papyrus for the Paleo example.
(You built one in the OCL lab, for example.)
Turn it into ecore like this:
- Create a new empty EMF project (File -> New -> Project -> Eclipse
Modeling Framework -> Empty EMF Project)
- Right click its model, create a new genmodel (new->other->Eclipse
Modeling Framework ->EMF generator model). The name of the genmodel doesn't
matter much provided it ends with .genmodel.
- When prompted, say that you're creating the ecore model from an
existing UML model, which is in your workspace; navigate to it and select
it. All the defaults will do. Generate only the RootElement package (not
the uml2.types - they should be referenced).
(If you run into problems, fix them as best you can. For example, my Papyrus
model had an operation getYes of Question and also an association end
called yes owned by Question; that choked the wizard because there was a
clash between the existing operation name and the one it wanted to create
from the property (roughly). In fact of course these two were not really in
conflict, but it couldn't know that... You can fix such things by renaming
or removing either the attribute or the operation in question. You may also
get strange results if your UML model includes anything other than a class
diagram - if you end up with an ecore model containing Interactions, just
delete them.)
- An ecore model will be created. Have a look at it. If you used a
Papyrus model that had OCL constraints in it, you'll see they've been
turned into comments, for example, and that there are operations whose
names contain Inv. This is interesting, have a look - but for today's
purposes it's probably best to edit those out of the ecore file. You should
end up with a very simple ecore file which (in the OclInEcore concrete
syntax) looks something like this:
import ecore : 'http://www.eclipse.org/emf/2002/Ecore' ;
package Paleo : Paleo = 'http:///Paleo.ecore'
{
abstract class Node
{
operation display();
attribute text : String[1];
}
class Question extends Node
{
attribute isInitial : Boolean[1];
property yes : Node[1];
property no : Node[1];
}
class Result extends Node
{
attribute paleo : Boolean[1];
}
}
If you did have OCL constraints and delete the resulting stuff from the
ecore model, you'll also want to get rid of the corresponding stuff from the genmodel. The
easiest way to do this is to generate a new genmodel from the edited ecore
model! Right-click the tidied ecore, pick New->Other->Eclipse Modeling
Framework->EMF Generator Model, and follow your nose.
Install EuGENia
2018 note: This is slightly more delicate than we'd like, and may
depend on what's currently installed in your Eclipse in ways I don't
totally understand. If these instructions don't work for you, ask.
- Install GMF Tooling using this update site:
http://download.eclipse.org/modeling/gmp/gmf-tooling/updates/releases/
(not the Examples or Experimental parts).
- Install Epsilon using this update site:
http://download.eclipse.org/epsilon/updates/ (I didn't explore
what is the minimum set of options we need - just let it install
everything, I suggest)
- To work around a problem, you need to install this specific
version of QVTo. It's a zip file; save it (maybe in /tmp), then install
it by clicking Add and then Archive in the Help -> Install new software
dialog, then navigating to it.
Don't worry about software being unsigned, restart when prompted. Be sure
to allow each software installation to finish before starting the next! I'm
not sure you really have to restart in between
each installation, but I
generally do.
Design your graphical DSML
Open your ecore file with the OclInEcore editor, i.e. the one you used in
the OCL lab. (The EuGENia documentation assumes you're using a different
presentation for ecore called Emfatic, but since you're already somewhat
familiar with OclInEcore - and since Emfatic has never worked well for me!
- you may as well use that; feel free to experiment if you wish.)
Add annotations to say how you want your abstract syntax represented in
the ecore model to be given graphical concrete syntax. These annotations
are themselves ecore elements; you're not creating an annotated ecore
model, but rather, a bigger ecore model, that includes annotations.
Now, what this means is that you're adding instances of EAnnotation to your
ecore model as children of the ecore elements you're modelling. See the
diagrams of the ecore metamodel here.
As you see, any EModelElement, i.e. essentially anything in your ecore
model, can be annotated. For example, you'll add an annotation to your
Question class, and the annotation will be a sibling of the
attributes/properties your Question class already has.
Since there are different concrete syntaxes for ecore, just how you add an
annotation depends on which one you're using! The EuGENia tutorial assumes
you're using Emfatic and uses one syntax; this lab assumes you're using
OclInEcore and so uses slightly different one.
For example,
class Question extends Node
{
annotation _'gmf.node'
(
label = 'text'
);
}
says that you want a Question to be represented by a node (as opposed to an
edge) in the diagrammatic presentation, and that you want it labelled by
the value of its text attribute (which it inherits from Node, if you're
using the version of the paleo metamodel shared before). Things to note
include:
- the underscore in front of the kind of annotation and the quotes round
it;
- round, not curly, brackets round the arguments to the annotation;
- the placement of semi-colons (though Eclipse will help you if you get
these wrong, thanks to OclInEcore itself being the concrete syntax for a
DSL namely ecore... it's turtles all the way down).
Just to try it out, add that one annotation to your ecore model, and let's
build the graphical editor.
Before this will work, there's one change you have to make: EuGENia insists
that one Class in your ecore model is designated the diagram. Add this
piece of magic to your ecore model (inside the package):
class DietDiagram
{
annotation _'gmf.diagram';
property nodes : Node[*|1] { ordered composes };
}
Next, select your ecore file in the package explorer, right-click,
and pick Eugenia -> Generate GMF editor from the menu.
Try out your DSML
Once you get the successful build message, you can run your editor using
the Run menu (at the top of your Eclipse), picking Run Configurations,
selecting Eclipse Application, going to the Arguments tab, and putting
-Xms512M -Xmx1024M in as VM arguments. (Ignore the EuGENia tutorial page's
instructions to include PermSize and MaxPermSize arguments: they don't
exist any more.)
2018 note You may get a Validation popup complaining about
org.apache.xmlrpc - you can ignore this.
A new Eclipse will appear. In it, create a new Project of type General.
Inside your new project, create a paleo diagram using File -> New ->
Example. At the top of the "Select a wizard" popup, you should see a wizard
with a name corresponding to your ecore package - pick that one!
You should get a palette that lets you create Questions. Have a go. You'll
see they come up rectangular, which isn't quite what we want, and there's
no way to add arrows between them yet. However, you can set their
properties, including using a drop-down menu to specify which question is
connected by yes or no links to which other question.
Having built a model and saved it, have a look in the Package Explorer at
the files that have been saved. You'll see there's one for the abstract
syntax representation of the model you just built, and one for the diagram
itself. Open each of these with a text editor, to see what's "really"
there.
Getting more serious
Having got the basic editor-building to work, close down that Eclipse and
go back to where you were editing your ecore file.
By reading the
EuGENia tutorial, bring your DSML closer to the form we looked at right
at the start of the course. As a reminder, here's that diagram:
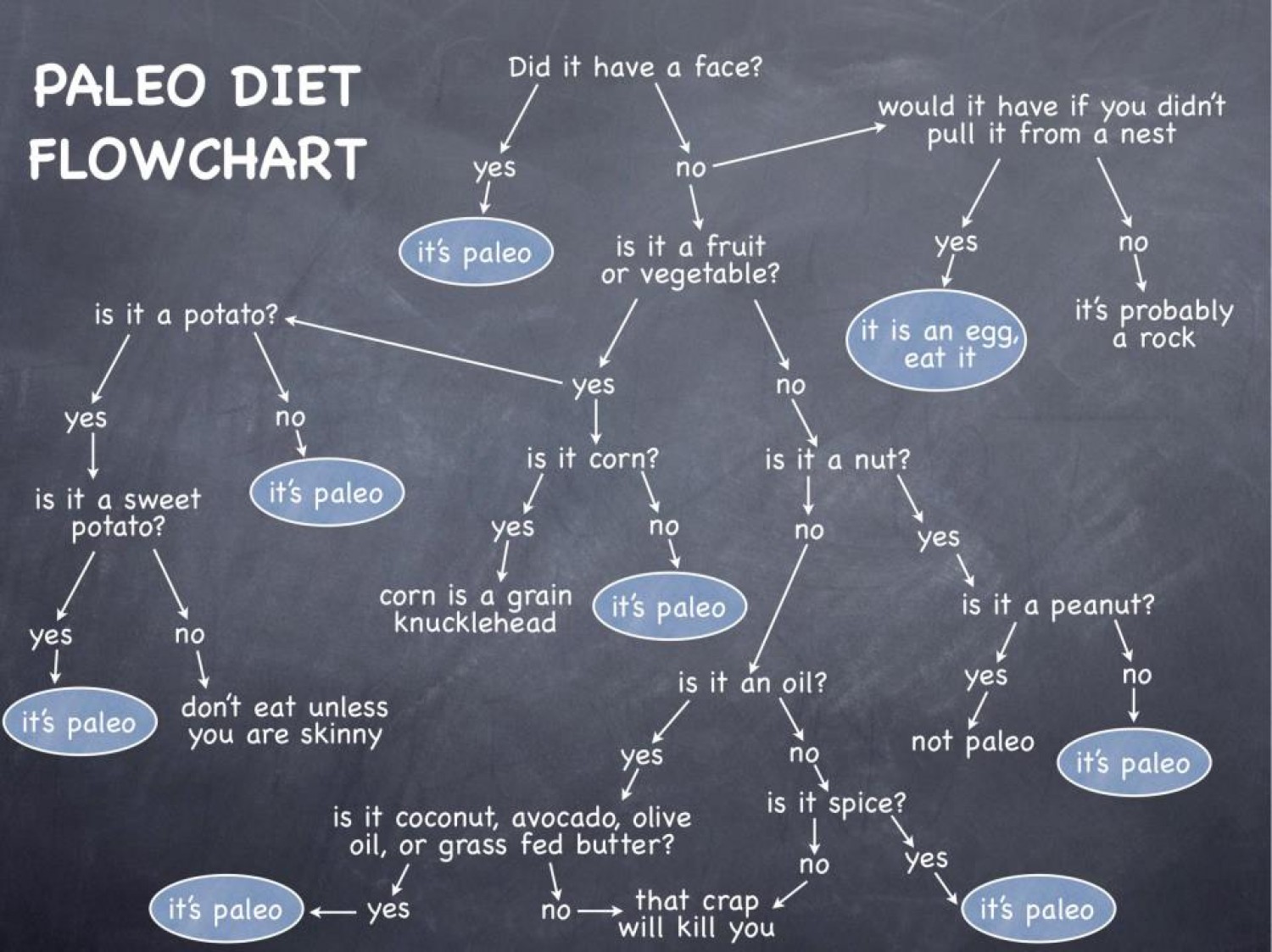
You probably won't be able to bring your DSML completely to this form - nor
should you, probably - but see if you can get it close enough that the
customer who asked you to build a tool for working with diets expressed
like this example they gave you would basically think you'd done the job.
2018 note: I haven't updated the notes below since 2016 - so some of
these problems may have gone away, and/or and others may have appeared.
Things you might want to know:
- I haven't found a way to set the background colour of the model, so my
version doesn't have the blackboard look of the original.
Perhaps that's a feature.
- For composite details to be given as part of an annotation, e.g.
border.color, the syntax is like what you saw for the type of annotation:
_'border.color' = ...
- The syntax for RGB colors is like '255,255,255'.
- I have not discovered what the syntax for giving a literal string is!
(That's why I use red and green edges in the screenshot below: bad
practice, because colourblindness.)
- To get edges to work, you will need to rejig your ecore metamodel,
because edges in a diagram have to be represented by things in the abstract
syntax. It's easy enough to do, but a little irritating, given that we
decided we didn't need to make arrows things in their own right to start
with. Could the change you have to make be automated? Challenge question:
automate it.
Here's a screenshot from part way through my effort. Can you get further?
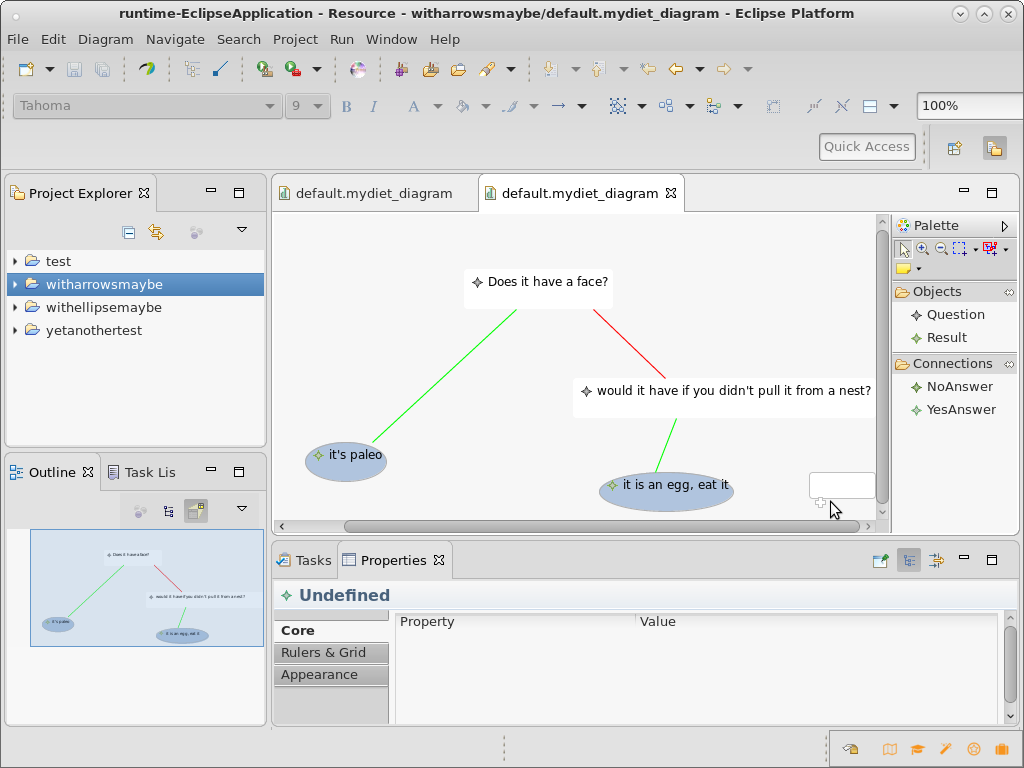
Further challenges for the brave and interested (optional!)
"For the brave" means that I don't know all the answers here...
Explore how you can do more than create a paleo-type diagram
Look at what else EuGENia can do for you, and see what you can work out
about what else could be done with an expression in your graphical DSML.
Presumably once your user has built and saved such an expression, i.e. a
model in your DSML - say, a flow diagram to tell whether a food is part of
a vegan diet - you'll want to use the model for something, e.g., save
effort in writing an Android app that can be used to check an individual
food. See how far you can get with understanding how the generated (or
generatable) code helps you. You may need to browse/google/read tutorials
and papers, rather than just look at the code.
Create a textual DSL from the ecore model
In principle, as well as creating a graphical syntax from your ecore file
as you did above, you can create a textual syntax from the ecore file,
using xtext and the "from existing ecore model" path. However, I ran into a
number of problems trying this, not all of which I solved.
- Right click your ecode model and pick "Register EPackages".
- Create a new xtext project, this time "from existing ecore model".
- Pick the genmodel corresponding to your ecore file (follow your
nose..).
- The wizard will attempt to create an xtext grammar from your ecore
model. It may not be completely successful (though I noticed it worked much
better in 2018 than in 2016!): if so, have a look at it and see if you can
fix it. (You can ignore the warning "The imported package is not on the
classpath of this project...")
- Compare the generated xtext grammar with the one you wrote by hand in
the xtext lab. The generated one is probably a bit
more complicated - see how much you can understand.
- As you did in the xtext lab, generate xtext artefacts and then try the
editor for the grammar. Try entering a diet in the syntax required by the
generated xtext grammar. Your mileage may vary, but for me this works:
Diet { nodes {
Question q1 {
text "does it have a face?"
yes r1
no r2
},
Result r1 {
text "eat it!"
},
Result r2 {
text "don't eat it!" }
}
}
Investigate the other components of the Epsilon toolkit...
... particularly model-to-model transformations in ETL, the Epsilon
Transformation Language. You'll find a paper describing how EuGENia is
implemented using model transformations
here.
This is the last lab of this presentation of SDM. There are many other
technologies we could have had "taster" labs on. For example, I would have
liked to fit in one or both of:
- ATL, probably the most widely-used model to model transformation
language (it's unidirectional, though).
- Triple Graph Grammars, TGGs, probably using eMOFLON.
You might like to explore these anyway (now or later!)
Note for students who may be looking for projects in future Among
other projects I'll be proposing will be one to develop more polished labs
suitable for students to use independently and/or in future presentations
of SDM. Such a lab might, for example, include better facilities for
students to check their own work (automated testing, rather than comparison
with a model answer). I'm thinking that ATL, eMOFLON, or Sirius might make
a good basis for one, for example, but I'm also open to having a lab on
[your favourite MDD technology] if you're interested in developing one. If
you're interested and would like to talk about it before the official list
comes out, drop me an email.
This page is maintained by
Perdita Stevens (
perdita@inf.ed.ac.uk)

