MLPR → Course Notes → w5b
PDF of this page
Previous: Backpropagation of Derivatives
Next: Netflix Prize
$$
\newcommand{\I}{\mathbb{I}}
\newcommand{\N}{\mathcal{N}}
\newcommand{\bb}{\mathbf{b}}
\newcommand{\bff}{\mathbf{f}}
\newcommand{\bh}{\mathbf{h}}
\newcommand{\bnu}{{\boldsymbol{\nu}}}
\newcommand{\bx}{\mathbf{x}}
\newcommand{\g}{\,|\,}
\newcommand{\te}{\!=\!}
\newcommand{\ttimes}{\!\times\!}
$$
Autoencoders and Principal Components Analysis (PCA)
One of the purposes of machine learning is to automatically learn how to use data, without writing code by hand. When we started the course with linear regression, we saw that we could represent complicated functions if we engineered features (or basis functions) by hand. Those functions can then be turned into “neural networks”, where — given enough labelled data — we can learn the features that are useful for classification.
For some data science tasks we can obtain a lot of unlabelled data: for example unannotated images, text, or user data. But the amount of relevant labelled data could be small. It can be useful to have features or basis functions that have been fitted on other tasks, which we can reuse. Then the number of parameters we need to fit for the current task is smaller.
Autoencoders
Autoencoders solve an unsupervised task: find a representation of feature vectors, without any labels. This representation might be useful for other tasks. An autoencoder is a neural network representing a vector-valued function, which when fitted well, approximately returns its input: \[
\bff(\bx) \approx \bx.
\] If we were allowed to set up the network arbitrarily, this function is easy to represent. For example with a single “weight matrix”: \[
\bff(\bx) = W\bx, \quad \text{with}~W=\I,~\text{the identity}.
\] Constraints are required to find an interesting representation that might be useful.
Dimensionality reduction: One constraint is to form a “bottleneck”. We use a neural network with a narrow hidden layer with \(K\ll D\) units: \[\begin{aligned}
\bh &= g^{(1)}(W^{(1)}\bx + \bb^{(1)})\\
\bff &= g^{(2)}(W^{(2)}\bh + \bb^{(2)}),
\end{aligned}\] where \(W^{(1)}\) is a \(K\ttimes D\) weight matrix, and the \(g\)’s are elementwise functions. If the function output manages to closely match its inputs, then we have a good lossy compressor. The network can compress \(D\) numbers down into \(K\) numbers, and then decode them again to nearly the original input.
One application of dimensionality reduction is visualization. When \(K\te2\) we can plot our transformed data as a 2-dimensional scatter-plot.
When an autoencoder works well, the transformed values \(\bh\) contain most of the information from the original input. We should therefore be able to use these transformed vectors as input to a classifier instead of our original data. It might then be possible to fit a classifier on smaller inputs using less labelled data.
Denoising and sparse autoencoders: Fitting an autoencoder with a large hidden layer can also be desirable. One reason is that it is easier to separate classes with a linear classifier if the inputs are high-dimensional. One regularization strategy is the denoising autoencoder: randomly set some of the features in the input to zero. Then if \(K\te D\) the best strategy is no longer for \(W^{(1)}\) to be the identity matrix. The hidden units should represent common conjunctions of multiple input features, so that missing ones can be reconstructed. Alternatively, sparse autoencoders only allow a small fraction of the \(K\) hidden units to take on non-zero values. That limitation forces the network to represent the input vector as a linear combination of a small number of different “sources”. A large number, \(K\), of different sources are possible, but only a few can be used for each example.
Principal Components Analysis (PCA)
When a linear autoencoder is used with the square loss function, then Principal Components Analysis (PCA) reduces the data in an equivalent way with two advantages. 1) Unlike the neural network approach, the fitted solution is unique and can be found using standard linear algebra operations. 2) The solutions for different \(K\) are nested, for a given datapoint, the feature \(h_1\) is the same in the solutions for all \(K\), \(h_2\) is the same for all \(K\!\ge\!2\), and so on.
In this note, PCA reduces the dimensionality of an \(N\ttimes D\) data matrix \(X\), by multiplying it by a \(D\ttimes K\) matrix \(V\). To keep the maths simpler, we will assume for the rest of this note that the \(D\)-dimensional feature vectors are zero mean. That means that the average row in \(X\) is the zero vector, in other words, that the average of the numbers in each column of \(X\) is zero. We centre the data to have this property before doing anything else.
We compute the covariance matrix of the points. As we’re assuming \(X\) is zero-mean, the covariance is \(\Sigma = \frac{1}{N}X^\top X\). Recalling the material on multivariate Gaussians, the covariance can be used to describe an ellipsoidal ball summarizing how the data is spread in space.
Some axes of the ellipsoid are often very short, and the data is “squashed” into a ball that only significantly extends in some directions. The eigenvectors of the covariance matrix point along the axes of the ellipsoid, and the longest axes are the eigenvectors with the largest eigenvalues. (You can take this statement on trust, or work through Q4b of tutorial 2 again, from which you might see it.)
PCA measures the position of a data-point along the most elongated axes of the ellipsoid by setting the columns of projection matrix \(V\) to the \(K\) eigenvectors of the covariance matrix associated with the largest \(K\) eigenvalues. Often (for zero mean data) we take eigenvectors of \(X^\top X\), dropping the factor of \(1/N\) in the covariance, which doesn’t change the directions of the eigenvectors.
Example Matlab/Octave code for zero-mean data:
% Find top K principal directions:
[V, E] = eig(X'*X);
[E,id] = sort(diag(E),1,'descend');
V = V(:, id(1:K)); % DxK
% Project to K-dims:}
X_kdim = X*V; % NxK
% Project back:
X_proj = X_kdim * V'; % NxD
An illustration for \(D\te2\) and \(K\te1\) is below:
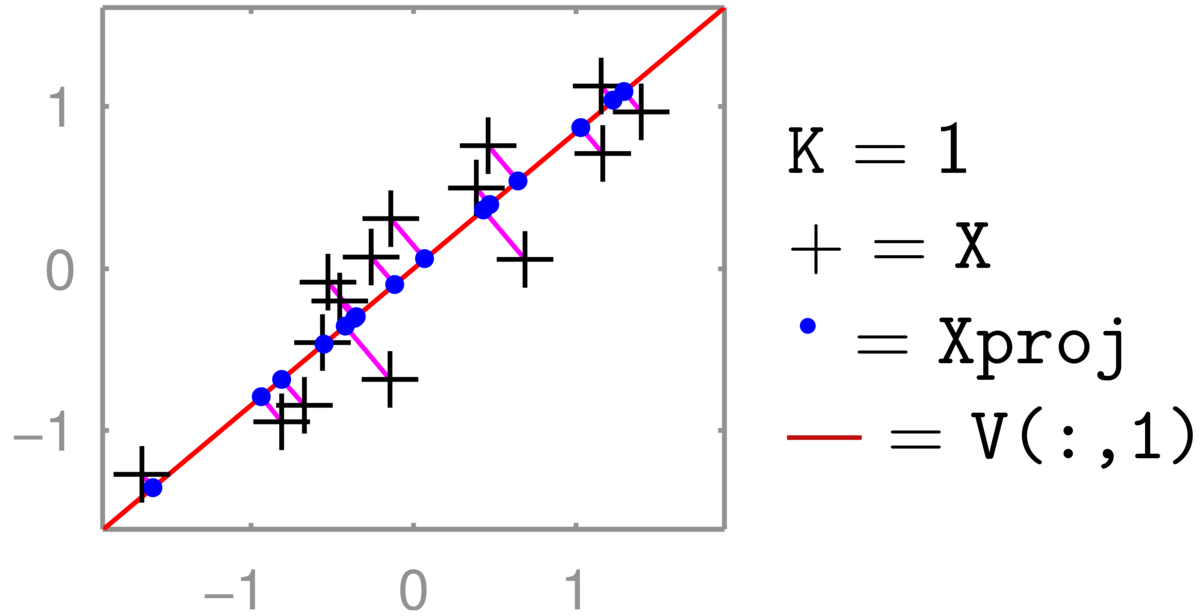
The two-dimensional coordinates of the pluses are reduced to one number, giving relative positions along the line that they have been projected onto (the principal component). Projecting back up to two dimensions gives the coordinates of this projected point in the full 2-dimensional space. However, the projected points \(XVV^\top\) are still constrained to lie along a one-dimensional line. (See also the related discussion in the pre-test.) The position along the second principal axis has been lost.
The eigenvectors of a covariance matrix are orthogonal, so if all the dimensions are kept, that is \(K\te D\), then \(VV^\top = \I\), and no information is lost.
We can replace a feature vector \(\bx\) with any projection \(A\bx\) in any model. The projection \(A\) could be generated at random (no fitting, so no risk of overfitting!). Alternatively the projection could be seen as \(K\ttimes D\) extra parameters, and fitted as part of the model (like in a neural network). PCA is a way to fit a sensible projection, but without fitting (and possibly overfitting) to a specific task.
PCA Examples
PCA is widely used, across many different types of data. It can give a quick first visualization of a dataset, or reduce the number of dimensions of a data matrix if overfitting or computational cost is a concern.
An example where we expect data to be largely controlled by a few numbers is body shape. The location of a point on a triangular mesh representing a human body is strongly constrained by the surrounding mesh-points, and could be accurately predicted with linear regression. PCA describes the principal ways in which variables can jointly change when moving away from the mean object. The principal components are often interpretable, and can be animated. Starting at a mean body mesh, one can move along each of the principal components, showing taller/shorter people, and then thinner/fatter people. The later principal components will correspond to more subtle, less interpretable combinations of features that covary.
A really neat PCA visualization was obtained by reducing the dimensionality of \(\approx 200,000\) features of people’s DNA to two dimensions (Novembre et al., 2008). The coordinates along the two principal axes closely correspond to a map of Europe showing where the people came from(!). (The people were carefully chosen.)
As is often the case with useful algorithms, we can choose how to put our data into them, and solve different tasks with the same code. Given an \(N\ttimes D\) matrix, we can run PCA to visualize the \(N\) rows. Or we can transpose the matrix and instead visualize the \(D\) columns. As an example, I took a binary \(S\ttimes C\) matrix \(M\) relating students and courses. \(M_{sc} \te 1\), if student \(s\) was taking course \(c\). In terms of these features, each course is a length-\(S\) vector, or each student is a length-\(C\) vector. We can reduce either of these sets of vectors to 2-dimensions and visualize them.
The 2D scatter plot of courses was somewhat interpretable:
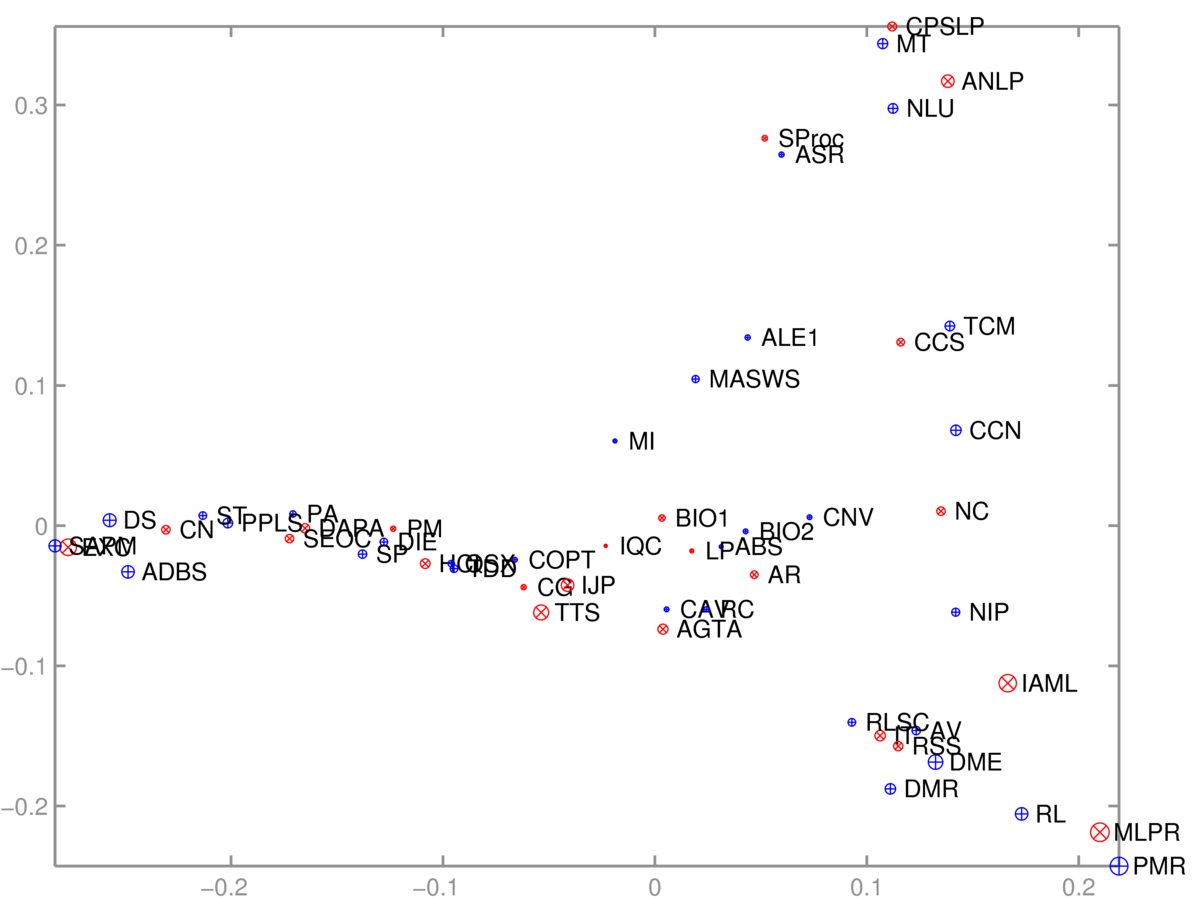
One axis (roughly) goes from computer-based applications of Informatics, through theory to broader applications. The other goes from cognitive/language applications down to machine learning. The algorithm had no labels, just which courses are taken together.
A scatter plot of students was less interpretable. I didn’t find obvious groups corresponding to the MSc specialisms offered in Informatics:
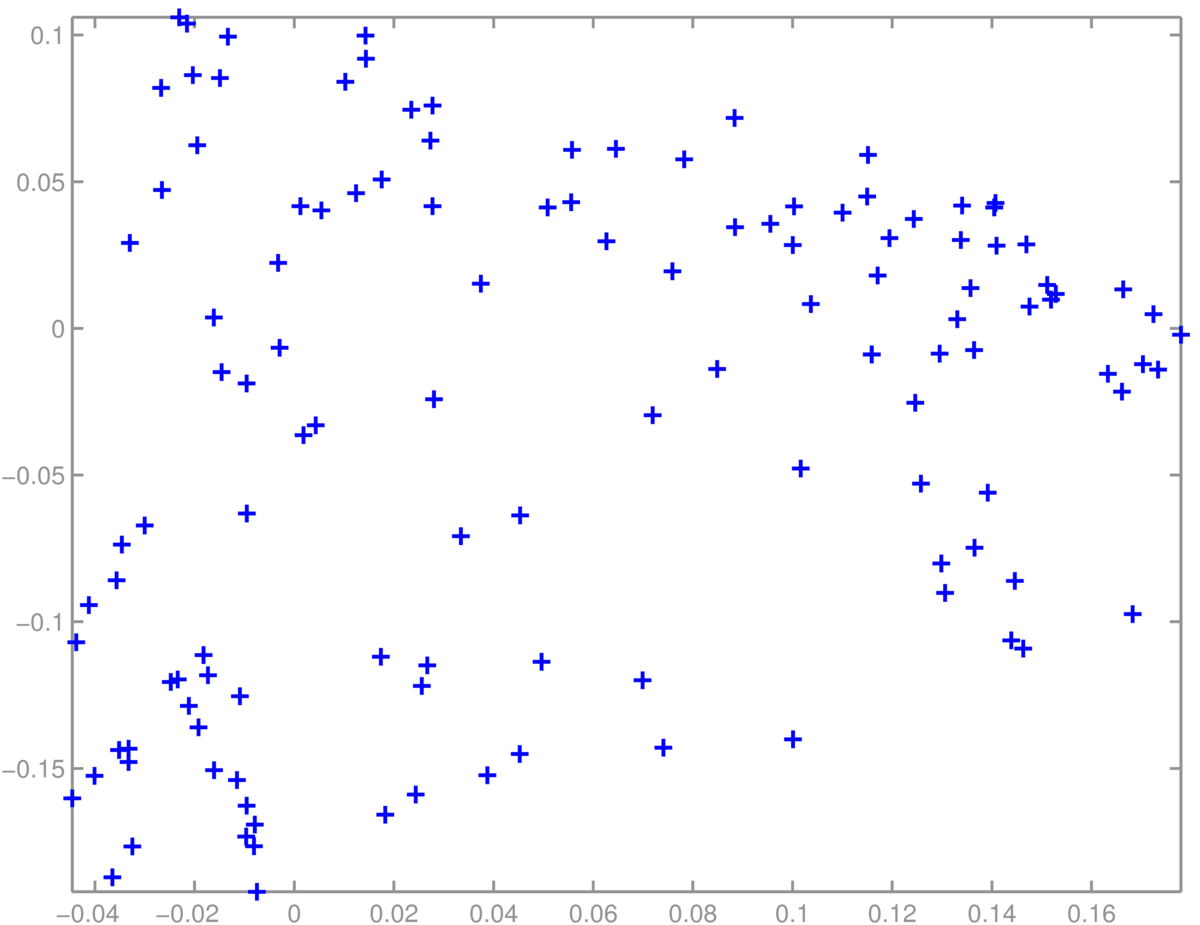
Finally, PCA doesn’t always work well. One of the papers that helped convince people that it was feasible to fit deep neural networks showed impressive results with non-linear deep autoencoders in cases where PCA worked poorly: Reducing the dimensionality of data with neural networks, Hinton and Salakhutdinov (2006). Science, Vol. 313. no. 5786, pp504–507, 28 July 2006. Available from http://www.cs.utoronto.ca/~hinton/papers.html
PCA and SVD
The truncated SVD view of PCA reflects the symmetry noted in the MSc course data example above: we can find a low-dimensional vector representing either the rows or columns of a matrix. SVD finds both at once.
Singular Value Decomposition (SVD) is a standard technique, available in most linear algebra packages. It factors a \(N\ttimes D\) matrix into a product of three matrices, \[
X \approx U S V^\top,
\] where \(U\) has size \(N\ttimes K\), \(S\) is a diagonal \(K\ttimes K\) matrix, and \(V^\top\) has size \(K\ttimes D\). The \(V\) matrix is the same as before, its columns (or the rows of \(V^\top\)) contain eigenvectors of \(X^\top X\). The columns of \(U\) contain eigenvectors of \(XX^\top\). The rows of \(U\) give a \(K\)-dimensional embedding of the rows of \(X\). The columns of \(V^\top\) (or the rows of \(V\)) give a \(K\)-dimensional embedding of the columns of \(X\).
Matlab/Octave demo:
% PCA via SVD, for zero-mean NxD matrix X
[U, S, V] = svd(X, 0);
U = U(:, 1:K); % NxK "datapoints" projected into K-dims
S = S(1:K, 1:K);
V = V(:, 1:K); % DxK "features" projected into K-dims
X_kdim = U*S;
X_proj = U*S*V';
A truncated SVD is known to be the best low-rank approximation of a matrix (as measured by square error). PCA is the linear dimensionality reduction method that minimizes the least squares error in the distortion if we project back to the original space: \(X\approx XVV^\top\).
Probabilistic versions of PCA
The simplest probabilistic model of \(D\)-dimensional feature vectors \(\bx\) that lie on a low-dimensional manifold, is to assume they’re Gaussian. The model assumes that a \(K\)-dimensional Gaussian variable was generated, \(\bnu \sim \N(\mathbf{0},I_K)\), and then projected up into \(D\)-dimensions, \(\bx = W\bnu\), where \(W\) is a \(D\ttimes K\) matrix. Under this model, \(\bx \sim \N(\mathbf{0}, WW^\top)\). The covariance is low rank, rank \(K\), because it only has \(K\) independent rows or columns. By the construction, we see that all vectors \(\bx\) generated from this model will lie exactly on a linear subspace of dimension \(K\).
A Gaussian with low-rank covariance isn’t able to explain real-world data, which won’t lie exactly on a linear subspace. Specifically the likelihood of such a model will be zero if any data-points lie outside the \(K\)-dimensional subspace. We can explain such deviations by assuming that spherical noise was added to the points from the model of the previous paragraph: \(\bx \sim \N(\mathbf{0}, WW^\top + \sigma_n^2I)\). This is the probabilistic PCA (PPCA) model. In the limit as \(\sigma_n^2\rightarrow 0\) the low-dimensional explanations of the data will be the same as PCA. But a more sensible model will result by setting non-zero \(\sigma_n^2\). PPCA is a special case of probabilistic Factor Analysis, which sets the noise to be an arbitrary diagonal covariance matrix.
Further reading
Murphy’s treatment of PCA: Section 12.2.1 p387–389, and Section 12.2.3 pp392–395.
Barber’s treatment starts in Section 5.2.
What you need to know
I’m not going to ask you to prove things about eigenvectors, or the relationships between eigendecompositions and SVD, or other deep technical details relating to PCA on the exam.
You should know how PCA can be done, because it’s useful. You should also be able to discuss why it may be better or worse than other dimensionality reduction or feature extraction methods you’re asked to consider. Multivariate Gaussians come up a lot in this course, so thinking about PPCA and Factor Analysis may be useful. Again you don’t need to memorize equations, but you could be given the details and then asked about them, just as with other Gaussian models we will cover.
Bonus note on matrix functions
Non-examinable!
In the past I’ve been asked: if \(X\) is a square symmetrical matrix, doesn’t the SVD of \(X\) give me the eigenvectors of \(X\)? Yes it does. That’s potentially confusing because above I said that it gives the eigenvectors of \(XX^\top\) and \(X^\top X\). For a square symmetrical matrix, the SVD therefore gives the eigenvectors of \(X^2\). These are in fact the same as the eigenvectors of \(X\), so there’s no contradiction.
\(X^2\) is the square function applied to the matrix \(X\). A way to apply a function to a covariance matrix is to decompose the matrix using the full SVD: \(X = USV^\top\), apply the function to the diagonal elements of \(S\) (in this case square the values), and then put the matrix back together again. The eigenvectors don’t change! It may interest you to know that other functions are applied to matrices in this way. For example the matrix exponential of a covariance matrix (expm in Matlab), equal to \(X+\frac{1}{2}X^2 + \frac{1}{3!}X^3 + \frac{1}{4!}X^4...\), can be computed by taking the SVD, exponentiating the singular values, and putting the matrix back together again. For a general square matrix, a function is applied to eigenvalues in an eigendecomposition \(X = U \Lambda U^{-1}\).
MLPR → Course Notes → w5b
PDF of this page
Previous: Backpropagation of Derivatives
Next: Netflix Prize
Notes by Iain Murray


